Antibodies: A Brief History
Antibodies are manifestations of disease, their presence indicating the early stages of illness and infection. They are ideal biomarkers with a wealth of information when profiled. As a matter of fact, they have been used for more than a century to detect disease, proving to be a robust reliable method of detecting and studying illness. The discoveries, developments, and innovations over the years applied to understanding antibodies have made it possible only recently to conduct immunoprofiling analyses. Each innovation, starting with the discovery of antibodies, collectively enhanced our ability to understand and treat disease. This introduction highlights these key breakthroughs.
For a complete timeline: Historical-Timeline-Milestones-in-Humoral-Immunity.pdf (sengenics.com)
The American Association of Immunologists posted a great interactive timeline The American Association of Immunologists – Interactive Timeline (aai.org)
Antibodies: The Antitoxin
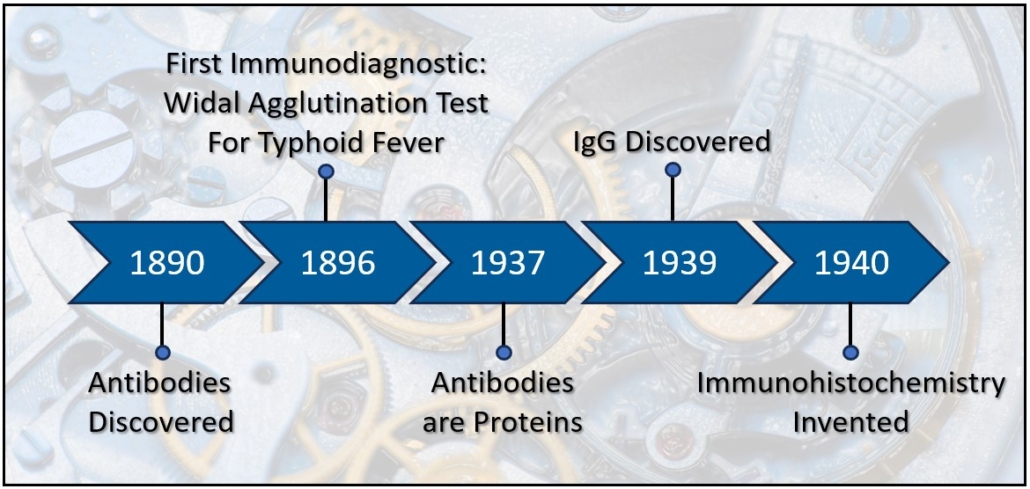
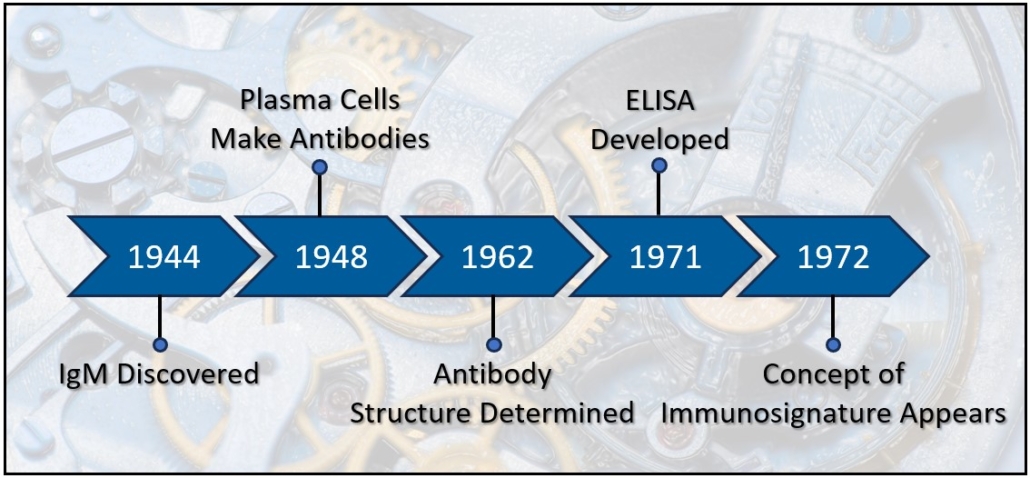
Antibodies were discovered in 1890 by Emil von Behring and Baron Kitasato Shibasaburō working together in Berlin using blood to find a cure for tetanus. They demonstrated that serum from a rabbit previously injected with tetanus prevents tetanus infection when transferred to naïve rabbits. Several important criteria emerged from these experiments that described the key principles of antibodies and the humoral immune system including 1) Protection from tetanus occurs cell-free, 2) The extravascular blood from immunized animals possess the capacity to neutralize tetanus, 3) The sera is stable and easily transferrable, 4) The protection is specific to the inoculant, 5) Neutralizing factor persists, even in the bodies of deceased animals [1]. In 1891, Paul Ehrlich used the word “Antikorper,” German for antibody, to differentiate the antitoxins of sera transfer from other methods of producing disease resistance such as variolization (nasal) [2-4]. The term antibody refers to the product produced by an inoculated organism that reacts against disease causing bacterial bodies. By 1901, antibody became the accepted term to describe the serum substance that transfers disease resistance from one organism to another. The material used to induce an antibody response was termed antigen in 1899 by the Hungarian microbiologist László Detre [5]. Immune studies of antibodies and soluble anti-toxins became known as humoral immunity and would eventually be linked to B Cells and autoimmunity. Other researchers of the early 20th century, such as Ilya Metchnikoff and Louis Pasteur, followed a different immunological research path. They observed phagocytosis of foreign bodies by host cells, i.e. cellular immunity, the basic activities of neutrophils and macrophages [4]. In 1939 the two camps joined together when Francine Sabin demonstrated that mononuclear cells both phagocytized foreign material and secreted an antibody-like globulin [6], and later, in 1948, Astrid Fagreus’ dissertation identified plasma cells as the source of antibodies [7, 8].
Antibodies: The Diagnostic
The key characteristics of antibodies described in von Behring’s and Shibasaburō‘s research suggested that antibodies could not only treat disease, but also identify disease. The specificity of the antibodies to disease was key. Fernand Widal developed the first immunodiagnostic test in 1896, a test for typhoid fever still used today in which positive patient antibodies cause agglutination of target infected cells [9, 10]. The antibody agglutination principle became applied to numerous diseases. Later, between 1901 and 1906, Jules Bordet developed a different antibody dependent diagnostic technique. He demonstrated that antibodies can interact with a “complement” to cause cell lysis, again specific to an infected cell. The complement fixation assay is basically a competitive inhibition between antibodies precoated on red blood cells and antibodies from a patient in a serum limited environment (the complement)[11]. The agglutination test and complement fixation tests were standards through the 20th century, highlighting the power of immunodiagnostics while also demonstrating some of the principles of how antibodies fight disease. As the 20th century progressed, more accurate and higher throughput diagnostic techniques surfaced including immunohistochemistry, enzyme-linked immunosorbent assays, imaging, and DNA and protein microarrays.
In 1940, Albert Coons and his colleagues took advantage of the known properties of antibodies and developed immunohistochemistry, a simpler more sensitive technique than the agglutination or complement fixation. Fluorescent tags are bound to the antibodies, and because antibodies are highly specific to antigen, the labeled antibody could be used to visualize proteins on or within cells from biopsies or in culture [12, 13]. Not only could this technique be used for diagnostics, but also to study the spatial distribution of proteins across tissues. This profound work launched antibodies as robust tools for diagnostics, histology and pathology throughout the 40’s and into modern times. The technique of indirect immunofluorescence became a mainstay in the study of proteomics. [14-16]. Although immunohistochemistry was a leap forward for immunodiagnostics and pathology, the technique initially lacked the throughput required for population sampling. Additionally, the assay was highly dependent upon the design and manufacture of antibodies.
Antibodies and Protein Microarrays
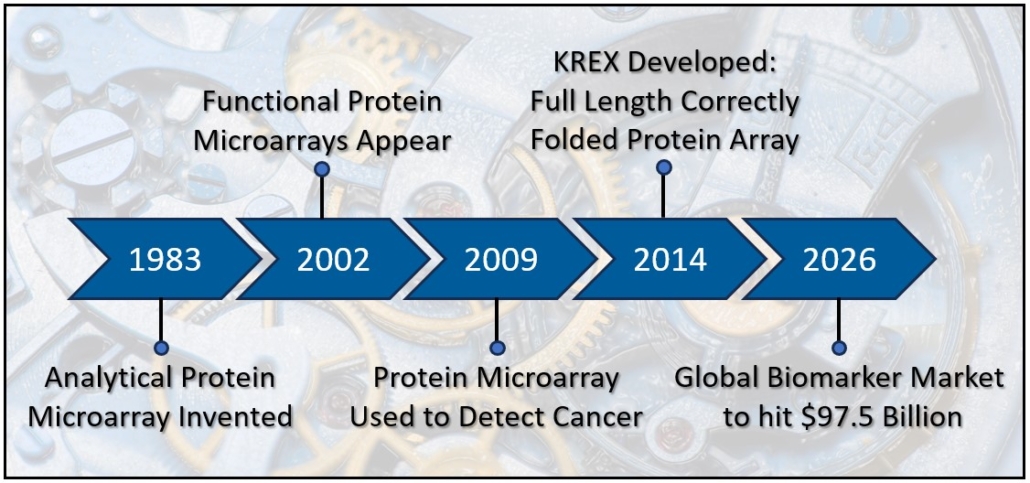
A number of advancements in immunodiagnostics happened in the 1970’s and 1980’s. ELISA appeared in 1971 [17] and offered numerous advantages to researchers including detection and quantification of proteins from multiple samples across a single microtiter plate. Later in the 1970’s, researchers applied species specific antibody fragments (Fc region) to a slide to immobilize cytotoxic T-cells in order to film their lysing activity. This study became the forerunner to modern protein microarray technology [18]. In 1983, Tse Wen Chang applied a series of different antibody spots across a slide in order to capture cells bearing different antigens [19], producing the first analytical protein microarray, in this case, an antigen capture microarray. Analytic microarrays and reverse phase microarrays were common throughout the 1980’s and 1990’s. In these assays, antibodies are used to detect proteins or proteins within tissue lysates. Proteins, in smaller amounts than required for ELISA, can be spotted across the slides, increasing throughput compared to ELISA or immunohistochemistry. By 2001, proteomics researchers had capitalized on recombinant DNA technology [20] and the human genome project. Protein libraries could now be synthesized using bacteria, yeast, or other hosts, and then collected and spot printed in rows and columns across a slide, up to several thousand proteins. Digital imaging technologies emerged that facilitated acquisition and quantification of the protein spots. By 2001, researchers could apply a donor serum to a protein microarray slide with thousands of proteins printed on its surface, and via indirect immunofluorescence and an array reader, identify hundreds of antibodies. Immunodiagnostics advanced from small scale assays based on antibody activities to large scale assays that identified the actual presence of hundreds of antibodies from a small serum sample, providing a much more complete and specific analysis of a patient. This individual profile, or immunoprofile, now has the power to not only identify disease, but also predict prognosis and identify patient subtypes.
Protein microarrays have become powerful tools for biomarker discovery. In the next blog, learn how protein microarray technology has evolved to provide high sensitivity, specificity and reproducibility.
References
1. Kantha, S.S., A centennial review; the 1890 tetanus antitoxin paper of von Behring and Kitasato and the related developments. Keio J Med, 1991. 40(1): p. 35-9.
2. Ehrlich, P., Croonian lecture.—On immunity with special reference to cell life. Proceedings of the Royal Society of London, 1900. 66(424-433): p. 424-448.
3. Winau, F., O. Westphal, and R. Winau, Paul Ehrlich–in search of the magic bullet. Microbes Infect, 2004. 6(8): p. 786-9.
4. Silverstein, A.M., Cellular versus humoral immunology: a century-long dispute. Nat Immunol, 2003. 4(5): p. 425-8.
5. Lindenmann, J., Origin of the terms ‘antibody’ and ‘antigen’. Scand J Immunol, 1984. 19(4): p. 281-5.
6. Sabin, F.R., Cellular Reactions to a Dye-Protein with a Concept of the Mechanism of Antibody Formation. J Exp Med, 1939. 70(1): p. 67-82.
7. Silverstein, A.M., Labeled antigens and antibodies: the evolution of magic markers and magic bullets. Nat Immunol, 2004. 5(12): p. 1211-7.
8. Norberg, R., G. Biberfeld, and H. Wigzell, Astrid Fagraeus, 1913–1997. Scandanavian Journal of Immunology, 2002. 47(1): p. 91.
9. Rose, N.R., The Concept of Immunodiagnosis, in Autoantibodies (Third Edition), Y. Shoenfeld, P.L. Meroni, and M.E. Gershwin, Editors. 2014, Elsevier. p. 3-10.
10. Olopoenia, L.A. and A.L. King, Widal agglutination test – 100 years later: still plagued by controversy. Postgrad Med J, 2000. 76(892): p. 80-4.
11. Bordet, J., Studies in Immunity. 1909, New York: Wiley and Sons.
12. Coons, A.H., H.J. Creech, and R.N. Jones, Immunological Properties of an Antibody Containing a Fluorescent Group. Proceedings of the Society for Experimental Biology and Medicine, 1941. 47(2): p. 200-202.
13. Coons, A.H. and M.H. Kaplan, Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J Exp Med, 1950. 91(1): p. 1-13.
14. Irure-Ventura, J. and M. Lopez-Hoyos, The Past, Present, and Future in Antinuclear Antibodies (ANA). Diagnostics (Basel), 2022. 12(3).
15. Blackburn, J.M., A. Shoko, and N. Beeton-Kempen, Miniaturized, microarray-based assays for chemical proteomic studies of protein function. Methods Mol Biol, 2012. 800: p. 133-62.
16. Duarte, J.S., J; Mulder, N; Blackburn, J., Protein Functional Microarrays: Design, Use and Bioinformatic Analysis in Cancer Biomarker Discovery and Quantitation, in Bioinformatics of Human Proteomics, X. Wang, Editor. 2013, Springer Science+Business Media Dordrecht. p. 39-74.
17. Aydin, S., A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides, 2015. 72: p. 4-15.
18. Rothstein, T.L., et al., Cytotoxic T lymphocyte sequential killing of immobilized allogeneic tumor target cells measured by time-lapse microcinematography. J Immunol, 1978. 121(5): p. 1652-6.
19. Chang, T.W., Binding of cells to matrixes of distinct antibodies coated on solid surface. J Immunol Methods, 1983. 65(1-2): p. 217-23.
20. Morrow, J.F., et al., Replication and transcription of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A, 1974. 71(5): p. 1743-7.