Proteomics Technology Spotlight – Functional Protein Microarrays
Proteins are the resulting expression of genetic information. They are the cellular workforce responsible for a myriad of physiological functions including providing structure, transporting nutrients, signaling with other cells, and regulating gene expression and translation of other proteins. Disease often impacts protein expression and function. Many neurodegenerative diseases, for example, produce aberrant protein tangles that disrupt physiology and lead to cognitive and behavioral impairment [1]. Understanding protein pathways is crucial to fully understanding and diagnosing disease and aiding in the development of therapeutics. Most drugs target proteins, including monoclonal antibody therapies [2]. Proteomics is the study and analysis of proteins and their interactions. There are many different tools available to study proteomics; however, functional protein microarrays are uniquely powerful, offering high throughput readouts of patient antibody profiles tools that provide rich individualized disease relevant data from small serum samples [3, 4].
In 2008, Sengenics introduced the patented KREX® protein folding technology to improve the current state of protein microarray fabrication methodology (Sengenics | KREX | Protein Folding Technology). The first functional protein microarrays run in the early 2000’s suffered from several factors that limited widespread usage. Cloning libraries were in their infancy, high throughput protein purification techniques were lacking, methods to print functional proteins needed development, and methods were needed to control inter- and intra- slide variability [5]. Different and inconsistent methodologies led to the propensity for inaccurate and irreproducible data. The Sengenics KREX technology addressed these issues brining functional protein microarrays to the forefront of proteomics technologies.
Functional Protein Microarrays
Sengenics specializes in functional protein microarrays. In a functional protein microarray, thousands of proteins are printed onto glass slides. The sample is most often patient serum, although urine, cerebrospinal fluid and other fluids have been used (Figure 1).
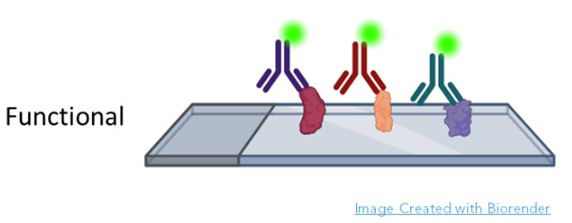
The metric is the visualization and quantification of each protein bound to cognate antibody (Figure 2). In many cases, these are autoantibodies because they react with human proteins printed on the slide. Autoantibodies are excellent biomarkers because they are limited in number, associated with inflammation, easy to obtain from blood, storable, and appear very early in disease. The AAg Atlas portal is dedicated to cataloging autoantibodies and contains comprehensive information about known autoantigens (AAgAtlas-FAQ (ncpsb.org.cn). Autoantibodies are among the first true signals of disease, and their identification not only indicates the presence of a health issue but may also be predictive of progression. For example, in patients at risk for Parkinson’s, Anuar et. al. discovered a panel of 22 autoantibodies present in patient serum up to four years prior to formal diagnosis [6]. Similarly, unique autoantibody repertoires have been discovered years before symptoms in cancer and autoimmune disease patients [7-10]. Since complex diseases such as Alzheimer’s and cancer can affect multiple protein pathways across various organs, autoantibodies make perfect biomarkers because the humoral immune systems surveys the entire body, demarcating potential hazards in all systems with the expression of disease specific autoantibodies. For example, DeMarshall et. al. found 50 serum autoantibodies associated with mild cognitive impairment, pre-Alzheimer’s diagnosis, cognate to antigens expressed within the brain and in many other parts of the body. These included functions related to cellular respiration, apoptosis, mRNA production, and voltage gated potassium channel formation [11]. Further, the panel highlighted proteins that had become pathogenic and pathways where treatment might alter the course of Alzheimer’s disease. Autoantibodies provide highly accurate early detection of the disease when organs are first affected. A functional protein microarray conducted with thousands of proteins at a time produces comprehensive results of diagnostic, prognostic, and therapeutic value. [6, 12-14].
Function Protein Microarrays with KREX® Technology
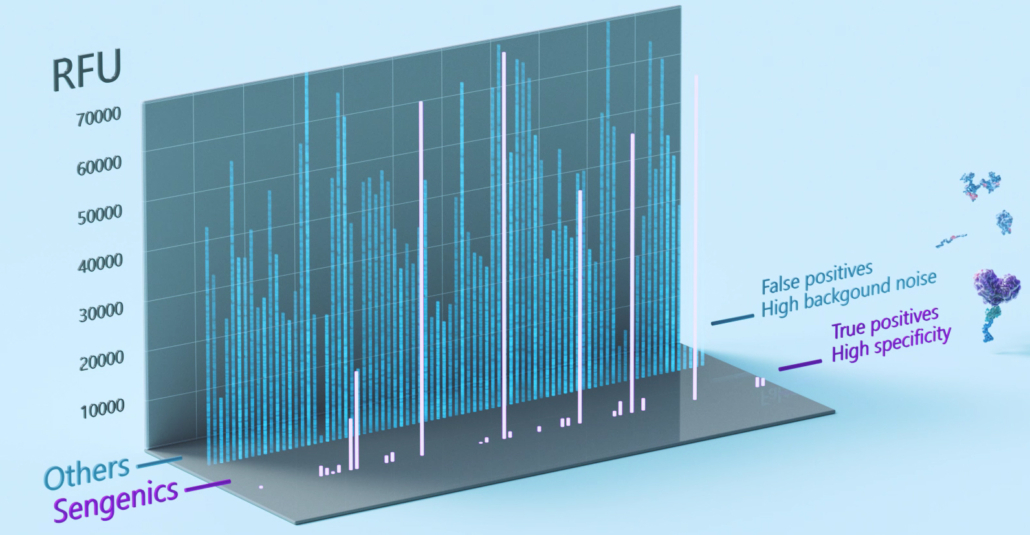
The Sengenics functional protein microarray contains carefully selected, biologically relevant, properly folded, and fully functional proteins. Attention to each of these details aids bioinformatics in a variety of ways. First, limiting the array to biologically relevant proteins results in meaningful, interpretable data by reducing noise from non-related proteins. This helps control false discovery rates and improve the likelihood of identifying biomarkers with high disease specificity and sensitivity (Figure 3). Second, because antibodies recognize specific shapes, proper folding of every chip protein is necessary to provide high antigen-antibody specificity and chip reproducibility [15-18]. To address this, Sengenics developed the patented KREX® technology.
With KREX® protein folding technology, a biotin carboxyl carrier molecule (BCCP) is coded in frame with each protein. A misfolded or fragmented protein results in BCCP misfolding, masking its biotin site and preventing it from binding to the streptavidin coated slide surface (Link to Movie). The biotin molecule binds with streptavidin coated onto a hydrogel microscope slide surface, adhering the protein in the same orientation for every printing. Hydrogel is an aqueous environment that helps the protein maintain its tertiary structure while also preventing the protein from flattening and bleeding into nearby spots. Last, The Sengenics i-Ome protein microarray is large enough for discovery and small enough to support smaller sample sizes (>30 recommended). Altogether, the array chips are highly consistent, highly specific, and highly reproducible making for a very cost-effective proteomics tool.
References
- Katsinelos, T., et al., The Role of Antibodies and Their Receptors in Protection Against Ordered Protein Assembly in Neurodegeneration. Front Immunol, 2019. 10: p. 1139.
- Santos, R., et al., A comprehensive map of molecular drug targets. Nat Rev Drug Discov, 2017. 16(1): p. 19-34.
- Sutandy, F.X.R., et al., Overview of Protein Microarrays. Current Protocols in Protein Science, 2013. 72(1): p. 27.1.1-27.1.16.
- Blackburn, J.M., A. Shoko, and N. Beeton-Kempen, Miniaturized, microarray-based assays for chemical proteomic studies of protein function. Methods Mol Biol, 2012. 800: p. 133-62.
- Bertone, P. and M. Snyder, Advances in functional protein microarray technology. FEBS J, 2005. 272(21): p. 5400-11.
- Anuar, N.D., et al., Identification of novel autoantibody signatures related to non-motor symptoms in individuals with high-risk of Parkinson’s Disease, using KREX-based functional protein microarrays. A cross-sectional study. 2022, Society For Neuroscience: Society For Neuroscience Conference 2023.
- Bizzaro, N., Autoantibodies as predictors of disease: the clinical and experimental evidence. Autoimmun Rev, 2007. 6(6): p. 325-33.
- Tan, E.M. and J. Zhang, Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev, 2008. 222: p. 328-40.
- Sexauer, D., E. Gray, and P. Zaenker, Tumour- associated autoantibodies as prognostic cancer biomarkers- a review. Autoimmun Rev, 2022. 21(4): p. 103041.
- Zaenker, P. and M.R. Ziman, Serologic autoantibodies as diagnostic cancer biomarkers–a review. Cancer Epidemiol Biomarkers Prev, 2013. 22(12): p. 2161-81.
- Demarshall, C.A., et al., Detection of Alzheimer’s disease at mild cognitive impairment and disease progression using autoantibodies as blood‐based biomarkers. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring, 2016. 3(1): p. 51-62.
- Aziz, F. and J. Blackburn, Autoantibody-Based Diagnostic Biomarkers:Technological Approaches to Discovery and Validation, in Autoantibodies and Cytokines, W.A. Khan, Editor. 2018, IntechOpen. p. 159-188.
- Da Gama Duarte, J., J.M. Peyper, and J.M. Blackburn, B cells and antibody production in melanoma. Mamm Genome, 2018. 29(11-12): p. 790-805.
- Immunoprofiling & the Humoral Immune System: A Guide to a New Era of Biomarker Discovery, K.S. Bittman and K. McGirr, Editors. 2023, Gen Publishing, Inc.: Online. p. 30.
- Duarte, J.G. and J.M. Blackburn, Advances in the development of human protein microarrays. Expert Rev Proteomics, 2017. 14(7): p. 627-641.
- Muro, Y., et al., Synthetic compound peptide simulating antigenicity of conformation-dependent autoepitope. J Biol Chem, 1994. 269(28): p. 18529-34.
- Barlow, D.J., M.S. Edwards, and J.M. Thornton, Continuous and discontinuous protein antigenic determinants. Nature, 1986. 322(6081): p. 747-8.
- Van Regenmortel, M.H.V., Mapping Epitope Structure and Activity: From One-Dimensional Prediction to Four-Dimensional Description of Antigenic Specificity. Methods, 1996. 9(3): p. 465-72.